Abstract
Background: Clonal heterogeneity of the AML blast cell population has been well documented using next generation sequencing (NGS) strategies. This heterogeneity extends to the presence of LSC fraction among the leukemic population: a dynamic compartment evolving to overcome cell selection pressure during disease progression and its treatment including allogeneic Hematopoietic Stem Cell Transplantation (HSCT). However, it remains difficult to discriminate between LSC and nHSC or other myeloid progenitors as MPP, LMPP or GMP. Here we present how to combine FLOWSom and KALUZA software to obtain robust identification among tumoral and normal progenitors. We compared the flowLSC Profile with stemness 17 genes score to identify patients with adverse outcome.
Methods: 11 AML patients from ALFA07-01 clinical trial diagnosed in Lyon University Hospital between 2009-2010 were included in this study. The quantification of CD34+CD38- fraction at diagnosis from total bulk leukemia was made as previously described. To better discriminate between LSC and nHSC we designed MFC panel based on 2-tube antibody combinations to identify the pattern of LSC in the CD34+CD38- cell compartment. CD34/CD38/CD45/CD117« backbone » was used in both tubes, completed by lineage defined markers CD7/CD56/CD19/CD13/CD33/ and/or LSC identified markers CD123 +/- CD90/CD45RA, CLL1, TIM3, CD97.
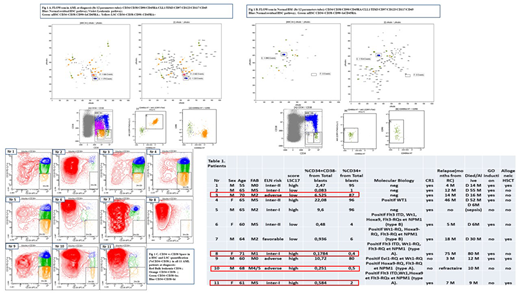
Results: As seeing in Table Nr 1, between 11 AML patients we observed a strong correlation identifying patients with adverse prognostic using previously published threshold by Flow (> 1% CD34+CD38- from bulk leukemia cells). 4 from 11 patients showed a discrepancy between GES 17 LSC and Flow CD34+CD38-: one from four patients (Nr 3) was classified as "high" by Flow (high CD34+ expression) and "low" by GES LSC 17 and three other patients (Nr 8, 10, 11) were classified inversely as "low" by Flow (<1% CD34+ expression) and "high" by GES LSC 17 scoring. Our preliminary data tend to show the relevance to establish a scoring system including the level of FlowLSC at diagnosis and GES LSC 17 to distinguish patients with high risk in patients with high CD34+ expression.
Conclusion: In summary, we confirmed that the presence of the LSC CD34+CD38- subpopulation representing more than 1% of total "bulk leukemic cells" at diagnosis could help to identify patients with a poor outcome. Using more accurate analysis as FLOWsom "neural network" system we showed the better discrimination between nHSC and LSC pathway with better separation between CD34+CD38- and CD34+CD38lo fractions and between nHSC (Fig 1A) and LSC (Fig 1B). This unsupervised system could allow us to improve and harmonize the analysis of AML LSC flow signature at diagnosis. A high LSC17 score probably reflects biological properties of LSCs that confer resistance to standard AML therapy. The previous analysis of the ALFA- 0701 trial data demonstrates the utility of the LSC17 score as a tool for patient selection. Despite heterogeneity and complexity of the AML LSC compartment, we should still use LSC quantification as a biomarker of chemosensitivity of AML (Fig 1C). For the patients with high-level of LSC other therapeutic modalities should be chosen to eradicate LSC using targeting immunotherapy before allograft. Monitoring of the LSC fraction should be useful in most clinical trials to overcome chemoresistance of LSC. These results should be confirmed in a prospectively larger cohort of patients and could be considered as useful complementary prognostic parameter for risk-stratification AML patients in future clinical trials.
Image: Fig1 A, B, C Table 1
Clinical Trial Registry: ALFA 0701
Conflict of Interest: No conflict
Key-Words: LSC, flow cytometry, AML, LSC17 Scoring
References:
Leukemic Stem Cell Frequency: A Strong Biomarker for Clinical Outcome in Acute Myeloid Leukemia : Monique Terwijn, et al, Gert J. Ossenkoppele1, Gerrit J. Schuurhuis ; PlosOne, 2014 Sep ;
Flow Cytometry to Estimate Leukemia Stem Cells in Primary Acute Myeloid Leukemia and in Patient-derived-xenografts, at Diagnosis and Follow Up. Boyer T, Gonzales F, Plesa A, et al, Roumier C, Preudhomme C, Cheok M; JoVE; March 2018;
A 17-gene stemness score for rapid determination of risk in acute leukaemia: Stanley W. K. Ng1, John E. Dick & Jean C. Y. Wang; Nature, 2017 Dec;
FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Van Gassen S, Callebaut B and Saeys Y (2017).
Dombret:CELLIPSE: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal